Introduction
1. Infrastructure Prerequisites
1.2 Panel/Exome
Resources for 1 VM:
-
64GB RAM
-
>= 40 cores
-
Filesystem: ext4/zfs
1.3 Genome
| gLeaves Instance | Worker1 | Worker2 | Worker3 | MongoDB | ElasticSearch | ClickHouse | RabbitMQ |
|---|---|---|---|---|---|---|---|
Backend + Frontend docker |
Annotations variants |
Index |
Recurrence |
Gene annotations |
Variant indexing |
Recurrence |
Message queue |
5GB RAM, 1 core, 500MB HDD |
16GB RAM, >=12 cores |
6GB RAM, 5 cores |
1GB RAM, 1 core |
4GB RAM, 1 core |
>=20GB RAM, 2 cores |
>=32GB RAM, >=16 cores |
512MB RAM, 1 core |
1.4 Volume Requirements
| Content | Volume | Path | Optional |
|---|---|---|---|
Docker images |
1.8GB |
/downloads/images |
No |
Test data |
40GB (20GB exome + 300MB panel) |
/downloads/test_dossiers |
Yes |
SpliceAI |
77GB |
/downloads/annotations/spliceAI |
No |
AlphaMissense |
2GB |
/downloads/annotations/AlphaMissense |
|
clinvar |
29MB |
/downloads/annotations/clinvar |
|
geneset |
21K |
/downloads/annotations/geneset |
|
mitomap |
121K |
/downloads/annotations/mitomap |
|
REVEL |
753MB |
/downloads/annotations/REVEL |
|
Nomad |
Coming soon |
/downloads/annotations/gnomADv4 |
Coming soon |
2. Software Architecture
![][](https://gleaves.laboratoire-seqoia.fr/docs/media/image2.png)
3. Deploying Docker Compose
3.1 Prerequisites
Ensure Docker is installed (minimum version with Docker Compose)
-
>= Docker version 19.03.15
-
>= docker-compose 1.29.2
It is necessary to load each of the provided Docker images.
docker load < rabbitmq_3-management.tar.gz docker load < minio.tar.gz docker load < clickhouse-server_23_3_1.tar.gz docker load < mongodb.tar.gz docker load < elasticsearch_6_8_14.tar.gz docker load < gleaves_3_5_33_5-1.tar.gz docker load < gleaves_worker_import_0_1_80-1.tar.gz docker load < gleaves_compute_rec_0_1_35-1.tar.gz docker load < gleaves_worker_index_0_1_35-1.tar.gz docker load < gleaves_ad_0_1_25-1.tar.gz
3.2 Configuration
-
gLeaves
Configuration of the SMTP server to send feedback emails (accessible in the UI) to a generic address receiving user feedback:
File: conf/gleaves/application.yml section [feedbackConfiguration]
smtpServer: smtp_hostname smtpPort: 25 authLogin: xxx.yyy@zzz.fr authPassword: xxx sendTo: xxx2.yyy2@zzz.fr
-
Team Configuration: File conf/gleaves/equipes.json
-
Workers:
Configuration of the SMTP server to send error/information emails, for the email address receiving issues. Files:
conf/workers/settings_import.toml conf/workers/settings_compute-rec.toml conf/workers/settings_index.toml
section [app]
email_rt_to="xxx.yyy@zzzfr" [email] server="smtp_hostname" port=25
2.3 Deployment & Access
Commands to execute for deployment: docker-compose up -d
All services are up:
user@server/gleaves_package docker-compose ps
Name/Port Command State
-------------------------------------------------------------------------------
gleaves_package_clickhouse_1 /entrypoint.sh Up 8123/tcp, 9000/tcp, 9009/tcp
gleaves_package_elasticsearch_1 /usr/local/bin/docker-entr ... Up 9200/tcp, 9300/tcp
gleaves_package_gleaves_1 /bin/sh -c java -jar $JAVA ... Up 0.0.0.0:443->6443/tcp
gleaves_package_gleaves_ad_1 /bin/sh -c JWT_SECRET=D7D4 ... Up 0.0.0.0:7443->7443/tcp
gleaves_package_minio_1 /usr/bin/docker-entrypoint ... Up 0.0.0.0:8900->8900/tcp, 0.0.0.0:9000->9000/tcp
gleaves_package_mongodb_1 docker-entrypoint.sh mongo ... Up 27017/tcp
gleaves_package_rabbitmq_1 docker-entrypoint.sh rabbi ... Up 15671/tcp, 15672/tcp, 15691/tcp, 15692/tcp, 25672/tcp, 4369/tcp, 5671/tcp, 0.0.0.0:5672->5672/tcp
gleaves_package_worker_compute_rec_1 /bin/bash -c RUST_LOG=info ...Up
gleaves_package_worker_import_1 /bin/bash -c RUST_LOG=info ... Up
gleaves_package_worker_index_1 /bin/bash -c RUST_LOG=info ... Up
-
The system is accessed via the following URLs:
gLeaves: https://server_hostname
gLeavesAD: https://server_hostname:7443
3. Operations
3.1 Accounts Creation
-
Admin Account
Default account created = admin/admin Create a new admin account with a password + delete the original admin account.
-
User Account
From the admin interface (see screenshot)
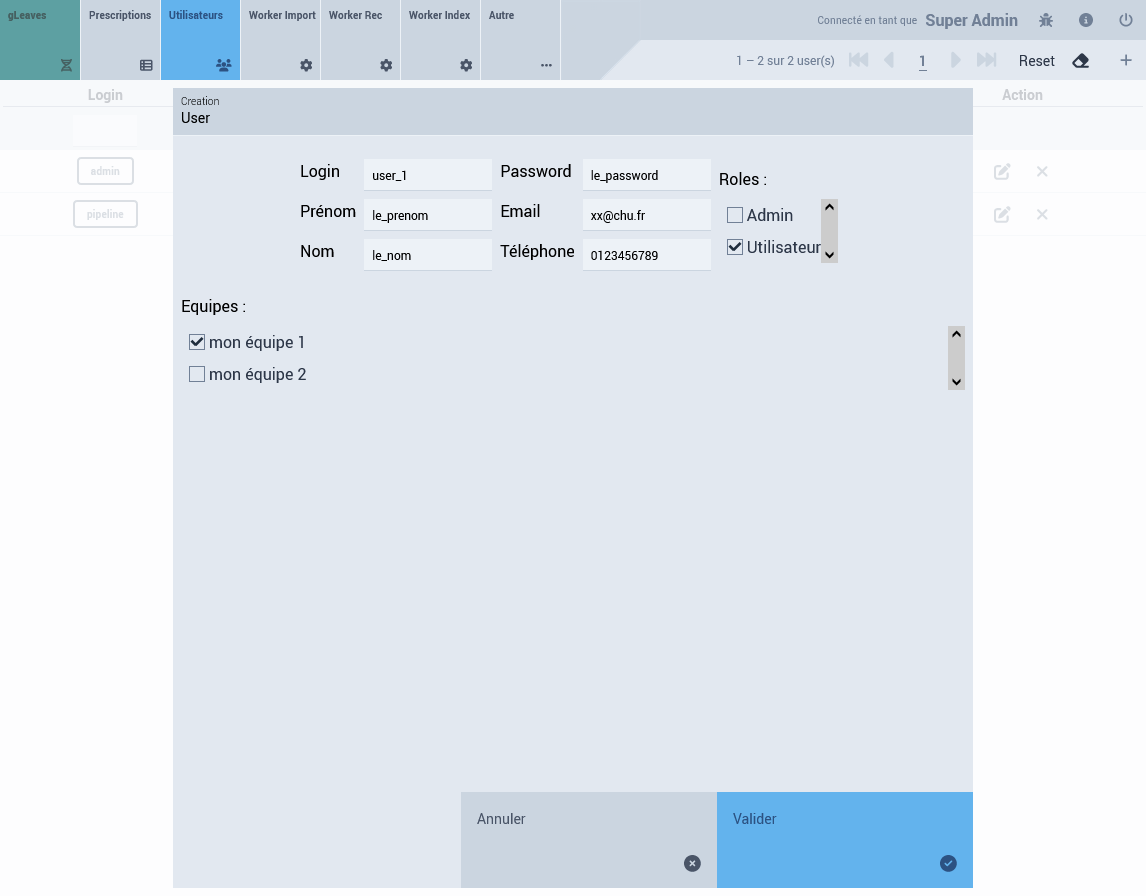
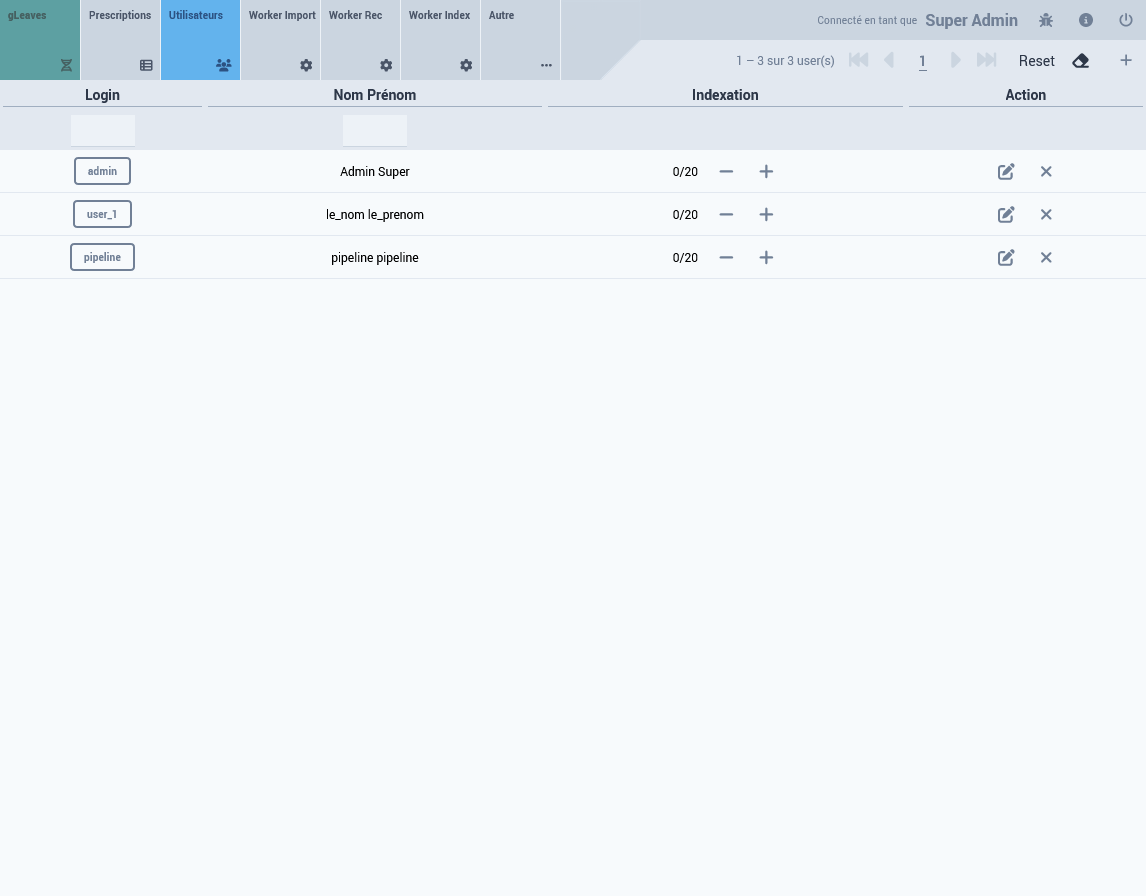
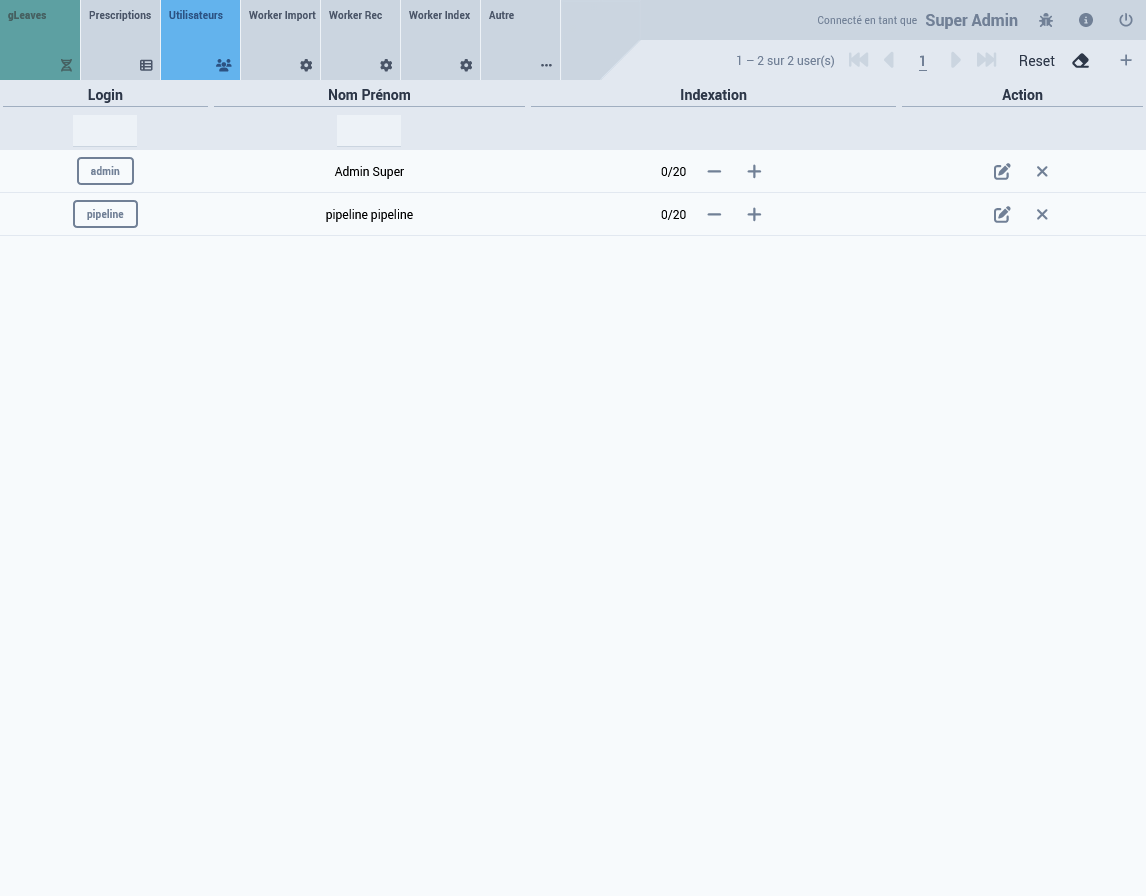
3.2 Data Insertion
3.2.1 VCF File Description
GT/AD/etc …
-
Path to example file
-
Annotation types
-
3.2.3 Annotations
3.2.3.1 Snpeff annotate
| Tool/Database | Version |
|---|---|
Genome |
GRCh38.92 |
snpEff |
4.3t |
#Download SnpEff databases
java -jar snpEff.jar download GRCh38.92java -Xmx7500m -Xms7500m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/snpEff.jar ann \
-c /data/annotations/snpeff_config_file/v4.3t/snpEff.config.seqoia \
-i vcf \
-o vcf \
-noInteraction \
-s {OUTPUT_SNPEFF_ANNOTATE.html} \
-dataDir /data/annotations/Human/GRCh38/index/snpEff/v4.3t \
GRCh38.92 \
{INPUT.vcf} 2>&1 1> \
{OUTPUT_SNPEFF_ANNOTATE.vcf}3.2.3.2 SnpSift DbNSFP
| Tool/Database | Version |
|---|---|
dbnsfp |
4.0a |
snpsift |
4.3t |
# Download dbNSFP database
wget http://dbnsfp.houstonbioinformatics.org/dbNSFPzip/dbNSFP4.0a.zip
# Uncompress
unzip dbNSFP4.0a.zip
# Create a single file version
# Compress using block-gzip algorithm
(zcat dbNSFP4.0a_variant.chr1.gz | head -n 1 - ; zcat dbNSFP4.0a_variant.chr* | grep -v "^#" ) | bgzip > dbNSFP4.0a.txt.gz
# Create tabix index
tabix -s 1 -b 2 -e 2 dbNSFP4.0a.txt.gzjava -Xmx7500m -Xms7500m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar dbnsfp \
-c /etc/snpeff/snpEff.config \
-db {DBNSFP_DATABASE.txt.gz} \
-v -collapse \
-f SIFT_score,SIFT_pred,Polyphen2_HDIV_score,Polyphen2_HDIV_pred,\
Polyphen2_HVAR_score,Polyphen2_HVAR_pred,LRT_score,LRT_pred,\
MutationTaster_score,MutationTaster_pred,FATHMM_score,FATHMM_pred,\
MetaSVM_score,MetaSVM_pred,MetaLR_score,MetaLR_pred,PROVEAN_score,\
PROVEAN_pred,CADD_raw,CADD_phred,GERP++_NR,GERP++_RS \
{INPUT_SNPEFF_ANNOTATE.vcf} 2>&1 1> \
{OUTPUT_SNPEFF_ANNOTATE.SNPSIFT_DBNSFP.vcf}3.2.3.4 SnpSift annotate
| Tool/Database | Version | Source |
|---|---|---|
snpsift |
4.3t |
|
kg(1000Genomes) |
phase3_20130502 |
|
gnomAD_exomes |
2.1.1 |
|
gnomAD_genomes |
3.0.0 |
|
dbsnp |
20180418 |
https://ftp.ncbi.nih.gov/snp/pre_build152/organisms/human_9606_b151_GRCh38p7/VCF/All_20180418.vcf.gz (splitted by chromosome, with standard chromosome names) |
cosmic_coding |
v89 (vcf split by chromosome, gzipped |
|
cosmic_noncoding |
v89 (vcf split by chromosome, gzipped) |
|
dbscsnv |
v1.1 |
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name kg_ \ +
-info EAS_AF,EUR_AF,AFR_AF,AMR_AF,SAS_AF \
{1000GENOME_DATABASECHROMOSOME.vcf.gz} \
{INPUT_SNPEFF_ANNOTATE.SNPSIFT_DBNSFP.vcf} | \
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name gnomAD_exomes_ \
-info non_par,AC0,InbreedingCoeff,PASS,RF,allele_type,variant_type,was_mixed,n_alt_alleles,AC_raw,AC,AC_female,AC_male,\
AC_popmax,AC_afr,AC_afr_female,AC_afr_male,AC_amr,AC_amr_female,AC_amr_male,AC_asj,AC_asj_female,AC_asj_male,AC_eas,\
AC_eas_female,AC_eas_male,AC_fin,AC_fin_female,AC_fin_male,AC_nfe,AC_nfe_female,AC_nfe_male,AC_oth,AC_oth_female,\
AC_oth_male,AC_sas,AC_sas_female,AC_sas_male,AF_raw,AF,AF_female,AF_male,AF_popmax,popmax,AF_afr,AF_afr_female,AF_afr_male,\
AF_amr,AF_amr_female,AF_amr_male,AF_asj,AF_asj_female,AF_asj_male,AF_eas,AF_eas_female,AF_eas_male,AF_fin,AF_fin_female,\
AF_fin_male,AF_nfe,AF_nfe_male,AF_nfe_female,AF_oth,AF_oth_female,AF_oth_male,AF_sas,AF_sas_female,AF_sas_male,AN_raw,AN,\
AN_female,AN_male,7AN_popmax,AN_afr,AN_afr_female,AN_afr_male,AN_amr,AN_amr_female,AN_amr_male,AN_asj,AN_asj_female,AN_asj_male,\
AN_eas,AN_eas_female,AN_eas_male,AN_fin,AN_fin_female,AN_fin_male,AN_nfe,AN_nfe_female,AN_nfe_male,AN_oth,AN_oth_female,AN_oth_male,\
AN_sas,AN_sas_female,AN_sas_male,controls_AC_raw,controls_AC,controls_AC_female,controls_AC_male,controls_AC_popmax,controls_AC_afr,\
controls_AC_afr_female,controls_AC_afr_male,controls_AC_amr,controls_AC_amr_female,controls_AC_amr_male,controls_AC_asj,\
controls_AC_asj_female,controls_AC_asj_male,controls_AC_eas,controls_AC_eas_female,controls_AC_eas_male,controls_AC_fin,\
controls_AC_fin_female,controls_AC_fin_male,controls_AC_nfe,controls_AC_nfe_female,controls_AC_nfe_male,controls_AC_oth,\
controls_AC_oth_female,controls_AC_oth_male,controls_AC_sas,controls_AC_sas_female,controls_AC_sas_male,controls_AF_raw,\
controls_AF,controls_AF_female,controls_AF_male,controls_AF_popmax,controls_popmax,controls_AF_afr,controls_AF_afr_female,\
controls_AF_afr_male,controls_AF_amr,controls_AF_amr_female,controls_AF_amr_male,controls_AF_asj,controls_AF_asj_female,\
controls_AF_asj_male,controls_AF_eas,controls_AF_eas_female,controls_AF_eas_male,controls_AF_fin,controls_AF_fin_female,\
controls_AF_fin_male,controls_AF_nfe,controls_AF_nfe_female,controls_AF_nfe_male,controls_AF_oth,controls_AF_oth_female,\
controls_AF_oth_male,controls_AF_sas,controls_AF_sas_female,controls_AF_sas_male,controls_AN_raw,controls_AN,controls_AN_female,\
controls_AN_male,controls_AN_popmax,controls_AN_afr,controls_AN_afr_female,controls_AN_afr_male,controls_AN_amr,controls_AN_amr_female,\
controls_AN_amr_male,controls_AN_asj,controls_AN_asj_female,controls_AN_asj_male,controls_AN_eas,controls_AN_eas_female,\
controls_AN_eas_male,controls_AN_fin,controls_AN_fin_female,controls_AN_fin_male,controls_AN_nfe,controls_AN_nfe_female,\
controls_AN_nfe_male,controls_AN_oth,controls_AN_oth_female,controls_AN_oth_male,controls_AN_sas,controls_AN_sas_female,\
controls_AN_sas_male,controls_nhomalt_raw,controls_nhomalt,controls_nhomalt_female,controls_nhomalt_male,controls_nhomalt_popmax,\
controls_nhomalt_afr,controls_nhomalt_afr_female,controls_nhomalt_afr_male,controls_nhomalt_amr,controls_nhomalt_amr_female,\
controls_nhomalt_amr_male,controls_nhomalt_asj,controls_nhomalt_asj_female,controls_nhomalt_asj_male,controls_nhomalt_eas,\
controls_nhomalt_eas_female,controls_nhomalt_eas_male,controls_nhomalt_fin,controls_nhomalt_fin_female,controls_nhomalt_fin_male,\
controls_nhomalt_nfe,controls_nhomalt_nfe_female,controls_nhomalt_nfe_male,controls_nhomalt_oth,controls_nhomalt_oth_female,\
controls_nhomalt_oth_male,controls_nhomalt_sas,controls_nhomalt_sas_female,controls_nhomalt_sas_male,nhomalt_raw,nhomalt,nhomalt_female,\
nhomalt_male,nhomalt_popmax,nhomalt_afr,nhomalt_afr_female,nhomalt_afr_male,nhomalt_amr,nhomalt_amr_female,nhomalt_amr_male,nhomalt_asj,\
nhomalt_asj_female,nhomalt_asj_male,nhomalt_eas,nhomalt_eas_female,nhomalt_eas_male,nhomalt_fin,nhomalt_fin_female,nhomalt_fin_male,\
nhomalt_nfe,nhomalt_nfe_female,nhomalt_nfe_male,nhomalt_oth,nhomalt_oth_female,nhomalt_oth_male,nhomalt_sas,nhomalt_sas_female,\
nhomalt_sas_male,non_cancer_AC_raw,non_cancer_AC,non_cancer_AC_female,non_cancer_AC_male,non_cancer_AC_popmax,non_cancer_AC_afr,\
non_cancer_AC_afr_female,non_cancer_AC_afr_male,non_cancer_AC_amr,non_cancer_AC_amr_female,non_cancer_AC_amr_male,non_cancer_AC_asj,\
non_cancer_AC_asj_female,non_cancer_AC_asj_male,non_cancer_AC_eas,non_cancer_AC_eas_female,non_cancer_AC_eas_male,non_cancer_AC_fin,\
non_cancer_AC_fin_female,non_cancer_AC_fin_male,non_cancer_AC_nfe,non_cancer_AC_nfe_female,non_cancer_AC_nfe_male,non_cancer_AC_oth,\
non_cancer_AC_oth_female,non_cancer_AC_oth_male,non_cancer_AC_sas,non_cancer_AC_sas_female,non_cancer_AC_sas_male,non_cancer_AF_raw,\
non_cancer_AF,non_cancer_AF_female,non_cancer_AF_male,non_cancer_AF_popmax,non_cancer_popmax,non_cancer_AF_afr,non_cancer_AF_afr_female,\
non_cancer_AF_afr_male,non_cancer_AF_amr,non_cancer_AF_amr_female,non_cancer_AF_amr_male,non_cancer_AF_asj,non_cancer_AF_asj_female,\
non_cancer_AF_asj_male,non_cancer_AF_eas,non_cancer_AF_eas_female,non_cancer_AF_eas_male,non_cancer_AF_fin,non_cancer_AF_fin_female,\
non_cancer_AF_fin_male,non_cancer_AF_nfe,non_cancer_AF_nfe_female,non_cancer_AF_nfe_male,non_cancer_AF_oth,non_cancer_AF_oth_female,\
non_cancer_AF_oth_male,non_cancer_AF_sas,non_cancer_AF_sas_female,non_cancer_AF_sas_male,non_cancer_AN_raw,non_cancer_AN,\
non_cancer_AN_female,non_cancer_AN_male,non_cancer_AN_popmax,non_cancer_AN_afr,non_cancer_AN_afr_female,non_cancer_AN_afr_male,\
non_cancer_AN_amr,non_cancer_AN_amr_female,non_cancer_AN_amr_male,non_cancer_AN_asj,non_cancer_AN_asj_female,non_cancer_AN_asj_male,\
non_cancer_AN_eas,non_cancer_AN_eas_female,non_cancer_AN_eas_male,non_cancer_AN_fin,non_cancer_AN_fin_female,non_cancer_AN_fin_male,\
non_cancer_AN_nfe,non_cancer_AN_nfe_female,non_cancer_AN_nfe_male,non_cancer_AN_oth,non_cancer_AN_oth_female,non_cancer_AN_oth_male,\
non_cancer_AN_sas,non_cancer_AN_sas_female,non_cancer_AN_sas_male,non_cancer_nhomalt_raw,non_cancer_nhomalt,non_cancer_nhomalt_female,\
non_cancer_nhomalt_male,non_cancer_nhomalt_popmax,non_cancer_nhomalt_afr,non_cancer_nhomalt_afr_female,non_cancer_nhomalt_afr_male,\
non_cancer_nhomalt_amr,non_cancer_nhomalt_amr_female,non_cancer_nhomalt_amr_male,non_cancer_nhomalt_asj,non_cancer_nhomalt_asj_female,\
non_cancer_nhomalt_asj_male,non_cancer_nhomalt_eas,non_cancer_nhomalt_eas_female,non_cancer_nhomalt_eas_male,non_cancer_nhomalt_fin,\
non_cancer_nhomalt_fin_female,non_cancer_nhomalt_fin_male,non_cancer_nhomalt_nfe,non_cancer_nhomalt_nfe_female,non_cancer_nhomalt_nfe_male,\
non_cancer_nhomalt_oth,non_cancer_nhomalt_oth_female,non_cancer_nhomalt_oth_male,non_cancer_nhomalt_sas,non_cancer_nhomalt_sas_female,\
non_cancer_nhomalt_sas_male,non_neuro_AC_raw,non_neuro_AC,non_neuro_AC_female,non_neuro_AC_male,non_neuro_AC_popmax,non_neuro_AC_afr,\
non_neuro_AC_afr_female,non_neuro_AC_afr_male,non_neuro_AC_amr,non_neuro_AC_amr_female,non_neuro_AC_amr_male,non_neuro_AC_asj,\
non_neuro_AC_asj_female,non_neuro_AC_asj_male,non_neuro_AC_eas,non_neuro_AC_eas_female,non_neuro_AC_eas_male,non_neuro_AC_fin,\
non_neuro_AC_fin_female,non_neuro_AC_fin_male,non_neuro_AC_nfe,non_neuro_AC_nfe_female,non_neuro_AC_nfe_male,non_neuro_AC_oth,\
non_neuro_AC_oth_female,non_neuro_AC_oth_male,non_neuro_AC_sas,non_neuro_AC_sas_female,non_neuro_AC_sas_male,non_neuro_AF_raw,\
non_neuro_AF,non_neuro_AF_female,non_neuro_AF_male,non_neuro_AF_popmax,non_neuro_popmax,non_neuro_AF_afr,non_neuro_AF_afr_female,\
non_neuro_AF_afr_male,non_neuro_AF_amr,non_neuro_AF_amr_female,non_neuro_AF_amr_male,non_neuro_AF_asj,non_neuro_AF_asj_female,\
non_neuro_AF_asj_male,non_neuro_AF_eas,non_neuro_AF_eas_female,non_neuro_AF_eas_male,non_neuro_AF_fin,non_neuro_AF_fin_female,\
non_neuro_AF_fin_male,non_neuro_AF_nfe,non_neuro_AF_nfe_female,non_neuro_AF_nfe_male,non_neuro_AF_oth,non_neuro_AF_oth_female,\
non_neuro_AF_oth_male,non_neuro_AF_sas,non_neuro_AF_sas_female,non_neuro_AF_sas_male,non_neuro_AN_raw,non_neuro_AN,non_neuro_AN_female,\
non_neuro_AN_male,non_neuro_AN_popmax,non_neuro_AN_afr,non_neuro_AN_afr_female,non_neuro_AN_afr_male,non_neuro_AN_amr,non_neuro_AN_amr_female,\
non_neuro_AN_amr_male,non_neuro_AN_asj,non_neuro_AN_asj_female,non_neuro_AN_asj_male,non_neuro_AN_eas,non_neuro_AN_eas_female,\
non_neuro_AN_eas_male,non_neuro_AN_fin,non_neuro_AN_fin_female,non_neuro_AN_fin_male,non_neuro_AN_nfe,non_neuro_AN_nfe_female,\
non_neuro_AN_nfe_male,non_neuro_AN_oth,non_neuro_AN_oth_female,non_neuro_AN_oth_male,non_neuro_AN_sas,non_neuro_AN_sas_female,\
non_neuro_AN_sas_male,non_neuro_nhomalt,non_neuro_nhomalt_female,non_neuro_nhomalt_male,non_neuro_nhomalt_popmax,non_neuro_nhomalt_raw,\
non_neuro_nhomalt_afr,non_neuro_nhomalt_afr_female,non_neuro_nhomalt_afr_male,non_neuro_nhomalt_amr,non_neuro_nhomalt_amr_female,\
non_neuro_nhomalt_amr_male,non_neuro_nhomalt_asj,non_neuro_nhomalt_asj_female,non_neuro_nhomalt_asj_male,non_neuro_nhomalt_eas,\
non_neuro_nhomalt_eas_female,non_neuro_nhomalt_eas_male,non_neuro_nhomalt_fin,non_neuro_nhomalt_fin_female,non_neuro_nhomalt_fin_male,\
non_neuro_nhomalt_nfe,non_neuro_nhomalt_nfe_female,non_neuro_nhomalt_nfe_male,non_neuro_nhomalt_oth,non_neuro_nhomalt_oth_female,\
non_neuro_nhomalt_oth_male,non_neuro_nhomalt_sas,non_neuro_nhomalt_sas_female,non_neuro_nhomalt_sas_male,non_topmed_AC,non_topmed_AC_female,\
non_topmed_AC_male,non_topmed_AC_popmax,non_topmed_AC_raw,non_topmed_AC_afr,non_topmed_AC_afr_female,non_topmed_AC_afr_male,\
non_topmed_AC_amr,non_topmed_AC_amr_female,non_topmed_AC_amr_male,non_topmed_AC_asj,non_topmed_AC_asj_female,non_topmed_AC_asj_male,\
non_topmed_AC_eas,non_topmed_AC_eas_female,non_topmed_AC_eas_male,non_topmed_AC_fin,non_topmed_AC_fin_female,non_topmed_AC_fin_male,\
non_topmed_AC_nfe,non_topmed_AC_nfe_female,non_topmed_AC_nfe_male,non_topmed_AC_oth,non_topmed_AC_oth_female,non_topmed_AC_oth_male,\
non_topmed_AC_sas,non_topmed_AC_sas_female,non_topmed_AC_sas_male,non_topmed_AF_raw,non_topmed_AF,non_topmed_AF_female,non_topmed_AF_male,\
non_topmed_AF_popmax,non_topmed_AF_afr,non_topmed_AF_afr_female,non_topmed_AF_afr_male,non_topmed_AF_amr,non_topmed_AF_amr_female,\
non_topmed_AF_amr_male,non_topmed_AF_asj,non_topmed_AF_asj_female,non_topmed_AF_asj_male,non_topmed_AF_eas,non_topmed_AF_eas_female,\
non_topmed_AF_eas_male,non_topmed_AF_fin,non_topmed_AF_fin_female,non_topmed_AF_fin_male,non_topmed_AF_nfe,non_topmed_AF_nfe_female,\
non_topmed_AF_nfe_male,non_topmed_AF_oth,non_topmed_AF_oth_female,non_topmed_AF_oth_male,non_topmed_AF_sas,non_topmed_AF_sas_female,\
non_topmed_AF_sas_male,non_topmed_AN_raw,non_topmed_AN,non_topmed_AN_female,non_topmed_AN_male,non_topmed_AN_popmax,non_topmed_popmax,\
non_topmed_AN_afr,non_topmed_AN_afr_female,non_topmed_AN_afr_male,non_topmed_AN_amr,non_topmed_AN_amr_female,non_topmed_AN_amr_male,\
non_topmed_AN_asj,non_topmed_AN_asj_female,non_topmed_AN_asj_male,non_topmed_AN_eas,non_topmed_AN_eas_female,non_topmed_AN_eas_male,\
non_topmed_AN_fin,non_topmed_AN_fin_female,non_topmed_AN_fin_male,non_topmed_AN_nfe,non_topmed_AN_nfe_female,non_topmed_AN_nfe_male,\
non_topmed_AN_oth,non_topmed_AN_oth_female,non_topmed_AN_oth_male,non_topmed_AN_sas,non_topmed_AN_sas_female,non_topmed_AN_sas_male,\
non_topmed_nhomalt_raw,non_topmed_nhomalt,non_topmed_nhomalt_female,non_topmed_nhomalt_male,non_topmed_nhomalt_popmax,non_topmed_nhomalt_afr,\
non_topmed_nhomalt_afr_female,non_topmed_nhomalt_afr_male,non_topmed_nhomalt_amr,non_topmed_nhomalt_amr_female,non_topmed_nhomalt_amr_male,\
non_topmed_nhomalt_asj,non_topmed_nhomalt_asj_female,non_topmed_nhomalt_asj_male,non_topmed_nhomalt_eas,non_topmed_nhomalt_eas_female,\
non_topmed_nhomalt_eas_male,non_topmed_nhomalt_fin,non_topmed_nhomalt_fin_female,non_topmed_nhomalt_fin_male,non_topmed_nhomalt_nfe,\
non_topmed_nhomalt_nfe_female,non_topmed_nhomalt_nfe_male,non_topmed_nhomalt_oth,non_topmed_nhomalt_oth_female,non_topmed_nhomalt_oth_male,\
non_topmed_nhomalt_sas,non_topmed_nhomalt_sas_female,non_topmed_nhomalt_sas_male,age_hist_het_bin_freq,age_hist_hom_bin_freq,dp_hist_all_bin_freq,\
dp_hist_alt_bin_freq \ +
{GNOMEAD_EXOME_DATABASECHROMOSOME} \
- | \
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name gnomAD_genomes_ \
-info non_par,AC0,AS_VQSR,InbreedingCoeff,PASS,variant_type,n_alt_alleles,AC_raw,AN_raw,AF_raw,nhomalt_raw,AC,AN,AF,nhomalt,AC_female,\
AN_female,AF_female,nhomalt_female,AC_male,AN_male,AF_male,nhomalt_male,AC_asj,AN_asj,AF_asj,nhomalt_asj,AC_asj_female,AN_asj_female,\
AF_asj_female,nhomalt_asj_female,AC_asj_male,AN_asj_male,AF_asj_male,nhomalt_asj_male,AC_fin,AN_fin,AF_fin,nhomalt_fin,AC_fin_female,\
AN_fin_female,AF_fin_female,nhomalt_fin_female,AC_fin_male,AN_fin_male,AF_fin_male,nhomalt_fin_male,AC_ami,AN_ami,AF_ami,nhomalt_ami,\
AC_ami_female,AN_ami_female,AF_ami_female,nhomalt_ami_female,AC_ami_male,AN_ami_male,AF_ami_male,nhomalt_ami_male,AC_oth,AN_oth,AF_oth,\
nhomalt_oth,AC_oth_female,AN_oth_female,AF_oth_female,nhomalt_oth_female,AC_oth_male,AN_oth_male,AF_oth_male,nhomalt_oth_male,AC_afr,\
AN_afr,AF_afr,nhomalt_afr,AC_afr_female,AN_afr_female,AF_afr_female,nhomalt_afr_female,AC_afr_male,AN_afr_male,AF_afr_male,nhomalt_afr_male,\
AC_eas,AN_eas,AF_eas,nhomalt_eas,AC_eas_female,AN_eas_female,AF_eas_female,nhomalt_eas_female,AC_eas_male,AN_eas_male,AF_eas_male,\
nhomalt_eas_male,AC_sas,AN_sas,AF_sas,nhomalt_sas,AC_sas_female,AN_sas_female,AF_sas_female,nhomalt_sas_female,AC_sas_male,AN_sas_male,\
AF_sas_male,nhomalt_sas_male,AC_nfe,AN_nfe,AF_nfe,nhomalt_nfe,AC_nfe_female,AN_nfe_female,AF_nfe_female,nhomalt_nfe_female,AC_nfe_male,\
AN_nfe_male,AF_nfe_male,nhomalt_nfe_male,AC_amr,AN_amr,AF_amr,nhomalt_amr,AC_amr_female,AN_amr_female,AF_amr_female,nhomalt_amr_female,\
AC_amr_male,AN_amr_male,AF_amr_male,nhomalt_amr_male \
{GNOMEAD_GENOME_DATABASECHROMOSOME} \
- | \
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name dbsnp_ \
-info RS \
{DBSNP_DATABASECHROMOSOME.vcf.gz}
- | \
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name cosmic_coding_ \
-info ID \
{COSMIC_CODING_DATABASECHROMOSOME} \
- | \
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name cosmic_noncoding_ \
-info ID \
{COSMIC_NONCODING_DATABASECHROMOSOME} \
- | \
java -Xmx1000m -Xms1000m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar annotate \
-c /etc/snpeff/snpEff.config -noDownload -noId -tabix \
-name dbscsnv_ \
-info ADA_SCORE,RF_SCORE \
{DBSCSNV_DATABASECHROMOSOME} \
- 2>&1 1> {OUTPUT_SNPEFF_ANNOTATE.SNPSIFT_DBNSFP.SNPSIFT_ANNOTATE.vcf}3.2.3.5 SnpSift phastcons
| Tool/Database | Version |
|---|---|
Genome |
GRCh38.92 |
snpsift |
4.3t |
Phastcons |
http://hgdownload.cse.ucsc.edu/goldenPath/hg38/phastCons100way/ |
java -Xmx7500m -Xms7500m -XX:ParallelGCThreads=1 -XX:+AggressiveHeap \
-jar /usr/share/java/SnpSift.jar phastcons \
-c /etc/snpeff/snpEff.config \
{PHASTCONS_DATABASE} \
{INPUT_SNPEFF_ANNOTATE.SNPSIFT_DBNSFP.SNPSIFT_ANNOTATE.vcf} 2>&1 \
1> {OUTPUT_SNPEFF_ANNOTATE.SNPSIFT_DBNSFP.SNPSIFT_ANNOTATE.SNPSIFT_PHASTCONS.vcf}3.2.4 Data Insertion
Command for test data insertion
cd send_commands ./gleaves_send_import ./settings.toml --index-if-p2 --with-cr-technique --json-path /test_dossiers/dossier_6A61/A00666_0012_WGS_MR_FS00505001_21042022.json